Replicating gsynth
Posted onIntroduction
Synthethic methods are a method of causal inference that seeks to combine traditional difference-in-difference types studies with time series cross sectional differences with factor analysis for uncontrolled/ unobserved measures. This method has been growing our of work initially from Abadie (Abadie, Diamond, and Hainmueller 2011a) and growing in importance/ research for state level policy analysis. It is interesting from the fact that it combines some elements of factor analysis to develop predictors and regression analysis to try to capture explained and unexplained variance.
Package
Method Assumptions
This methodology makes several critical assumptions.
- The model takes the functional form of
- Strict exogeneity (e.g. the error terms are indepdent to D, X, and f)
- Weak serial dependence of the error terms
- Regularity of conditions
The method then uses bootstrapping for confidence intervals.
There is some basic data available in the package
The data that will be initially used will be the simdata dataset
| id | time | Y | D | X1 | X2 | eff | error | mu | alpha | xi | F1 | L1 | F2 | L2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 116 | 1 | 4.96 | 0 | 0.75 | 0.23 | 0.00 | -2.60 | 5 | -0.36 | 1.13 | 0.25 | 1.39 | 0.01 | -0.18 |
| 114 | 29 | 16.90 | 0 | 0.87 | 2.83 | 8.96 | 0.50 | 5 | -1.53 | 3.24 | 0.18 | 0.25 | -0.38 | -0.78 |
| 101 | 10 | 1.44 | 0 | 1.52 | -0.55 | 0.00 | -0.74 | 5 | -0.06 | -1.60 | 0.22 | -0.04 | 1.15 | -0.88 |
| 125 | 6 | 15.78 | 0 | 2.20 | 2.91 | 0.00 | -0.99 | 5 | -0.65 | 0.70 | 1.52 | 0.54 | 0.33 | -0.09 |
| 129 | 11 | 9.63 | 0 | 0.14 | 1.34 | 0.00 | 0.19 | 5 | 1.62 | -0.53 | 0.38 | -0.73 | 0.99 | -0.51 |
| 101 | 12 | 5.47 | 0 | 1.43 | 0.33 | 0.00 | 0.04 | 5 | -0.06 | -1.46 | -0.50 | -0.04 | 0.55 | -0.88 |
| 119 | 9 | 10.94 | 0 | 2.65 | 2.17 | 0.00 | -0.09 | 5 | -1.20 | -1.51 | 0.12 | -0.60 | -0.33 | 1.02 |
| 134 | 18 | 13.71 | 0 | 1.72 | 2.28 | 0.00 | -0.96 | 5 | -0.81 | 0.33 | 0.05 | 1.02 | 1.53 | 1.00 |
| 140 | 4 | 7.83 | 0 | 2.38 | -0.18 | 0.00 | -0.21 | 5 | 1.28 | 1.91 | 1.37 | -0.93 | 0.64 | -1.13 |
| 133 | 25 | 12.05 | 0 | 1.08 | 1.97 | 3.45 | 0.30 | 5 | 0.32 | -0.37 | -0.69 | 0.66 | -0.95 | -0.29 |
| 109 | 15 | 17.43 | 0 | 2.98 | 2.98 | 0.00 | -0.56 | 5 | 0.69 | -1.29 | -1.07 | 0.18 | 1.36 | 1.37 |
| 104 | 23 | 21.17 | 1 | 2.60 | 2.27 | 3.52 | 0.07 | 5 | 1.34 | -0.37 | 1.01 | 2.02 | -0.25 | -0.62 |
| 128 | 30 | 4.79 | 0 | 2.31 | -0.50 | 8.95 | -2.55 | 5 | 1.63 | -0.42 | -0.14 | 0.33 | 0.92 | 0.39 |
| 119 | 8 | 6.02 | 0 | 0.83 | 0.99 | 0.00 | -0.09 | 5 | -1.20 | -1.57 | 0.58 | -0.60 | 0.44 | 1.02 |
| 122 | 7 | 8.50 | 0 | 0.71 | 1.23 | 0.00 | -0.66 | 5 | 0.56 | -0.26 | -1.55 | 0.67 | 1.10 | 0.45 |
| 122 | 2 | 9.68 | 0 | 2.00 | 1.01 | 0.00 | 0.39 | 5 | 0.56 | -1.46 | -0.03 | 0.67 | 0.39 | 0.45 |
| 104 | 25 | 9.31 | 1 | 0.39 | -0.53 | 5.33 | 0.02 | 5 | 1.34 | -0.37 | -0.69 | 2.02 | -0.95 | -0.62 |
| 109 | 16 | 12.81 | 0 | -1.41 | 2.24 | 0.00 | 1.78 | 5 | 0.69 | 0.79 | 0.30 | 0.18 | -0.60 | 1.37 |
| 143 | 8 | 6.43 | 0 | -0.36 | 0.90 | 0.00 | 0.48 | 5 | 0.60 | -1.57 | 0.58 | -0.30 | 0.44 | -0.54 |
| 134 | 27 | 2.32 | 0 | -0.13 | 0.36 | 8.13 | -0.71 | 5 | -0.81 | -1.05 | -0.28 | 1.02 | -0.78 | 1.00 |
| 125 | 16 | 3.00 | 0 | 0.66 | -0.67 | 0.00 | -1.01 | 5 | -0.65 | 0.79 | 0.30 | 0.54 | -0.60 | -0.09 |
| 130 | 17 | -7.68 | 0 | -0.10 | -3.48 | 0.00 | -0.19 | 5 | 0.79 | 0.77 | 0.45 | -1.22 | 2.19 | -1.35 |
| 129 | 10 | 7.47 | 0 | 1.15 | 0.75 | 0.00 | -0.21 | 5 | 1.62 | -1.60 | 0.22 | -0.73 | 1.15 | -0.51 |
| 132 | 29 | 10.77 | 0 | -0.02 | 1.23 | 11.04 | -0.19 | 5 | -0.96 | 3.24 | 0.18 | 1.39 | -0.38 | 0.58 |
| 143 | 29 | 13.15 | 0 | 2.11 | 0.24 | 8.82 | 1.33 | 5 | 0.60 | 3.24 | 0.18 | -0.30 | -0.38 | -0.54 |
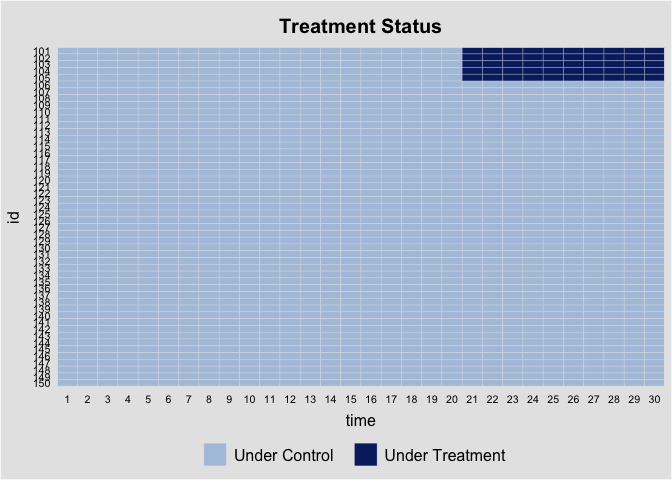
It can be examined via the panelView package:
Modeling
out <-
Cross-validating ...
r = 0; sigma2 = 1.84865; IC = 1.02023; PC = 1.74458; MSPE = 2.37280
r = 1; sigma2 = 1.51541; IC = 1.20588; PC = 1.99818; MSPE = 1.71743
r = 2; sigma2 = 0.99737; IC = 1.16130; PC = 1.69046; MSPE = 1.14540*
r = 3; sigma2 = 0.94664; IC = 1.47216; PC = 1.96215; MSPE = 1.15032
r = 4; sigma2 = 0.89411; IC = 1.76745; PC = 2.19241; MSPE = 1.21397
r = 5; sigma2 = 0.85060; IC = 2.05928; PC = 2.40964; MSPE = 1.23876
r* = 2
Simulating errors .............
Bootstrapping ...
..........
Call:
gsynth.formula(formula = Y ~ D + X1 + X2, data = simdata, index = c("id",
"time"), force = "two-way", r = c(0, 5), CV = TRUE, se = TRUE,
nboots = 1000, inference = "parametric", parallel = FALSE)
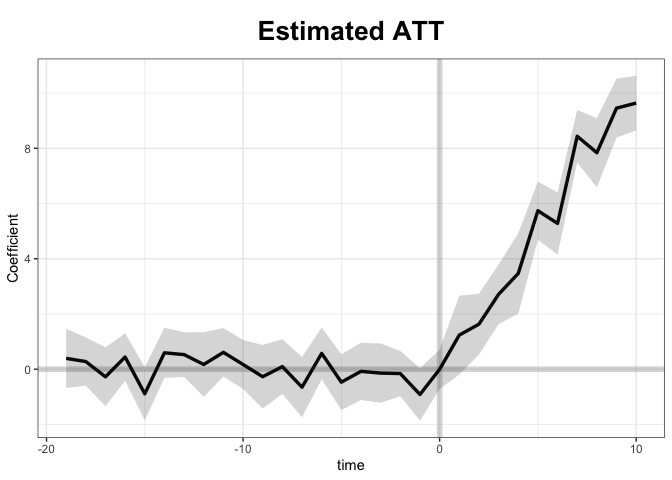
Average Treatment Effect on the Treated:
Estimate S.E. CI.lower CI.upper p.value
ATT.avg 5.544 0.262 5.03 6.057 0
~ by Period (including Pre-treatment Periods):
ATT S.E. CI.lower CI.upper p.value n.Treated
-19 0.392160 0.5468 -0.6796 1.46396 4.733e-01 0
-18 0.276548 0.4461 -0.5979 1.15098 5.354e-01 0
-17 -0.275393 0.5428 -1.3392 0.78841 6.119e-01 0
-16 0.441201 0.4389 -0.4191 1.30150 3.148e-01 0
-15 -0.889595 0.4914 -1.8527 0.07348 7.023e-02 0
-14 0.593891 0.4633 -0.3142 1.50199 1.999e-01 0
-13 0.528749 0.4132 -0.2811 1.33860 2.007e-01 0
-12 0.171569 0.5975 -0.9994 1.34258 7.740e-01 0
-11 0.610832 0.4474 -0.2661 1.48774 1.722e-01 0
-10 0.170597 0.4516 -0.7146 1.05581 7.056e-01 0
-9 -0.271892 0.5871 -1.4226 0.87886 6.433e-01 0
-8 0.094843 0.5037 -0.8924 1.08204 8.506e-01 0
-7 -0.651976 0.5567 -1.7430 0.43904 2.415e-01 0
-6 0.573686 0.4783 -0.3638 1.51113 2.304e-01 0
-5 -0.469686 0.5150 -1.4792 0.53978 3.618e-01 0
-4 -0.077766 0.5287 -1.1140 0.95850 8.831e-01 0
-3 -0.141785 0.5473 -1.2144 0.93085 7.956e-01 0
-2 -0.157100 0.4186 -0.9776 0.66341 7.075e-01 0
-1 -0.915575 0.4856 -1.8673 0.03616 5.936e-02 0
0 -0.003309 0.3638 -0.7164 0.70975 9.927e-01 0
1 1.235962 0.7289 -0.1927 2.66458 8.995e-02 5
2 1.630264 0.5615 0.5297 2.73079 3.691e-03 5
3 2.712178 0.5508 1.6326 3.79178 8.487e-07 5
4 3.466758 0.7437 2.0092 4.92431 3.136e-06 5
5 5.740132 0.5367 4.6882 6.79203 0.000e+00 5
6 5.280035 0.5741 4.1548 6.40529 0.000e+00 5
7 8.436485 0.4843 7.4874 9.38561 0.000e+00 5
8 7.839902 0.6408 6.5839 9.09592 0.000e+00 5
9 9.455115 0.5450 8.3869 10.52329 0.000e+00 5
10 9.638509 0.5035 8.6517 10.62527 0.000e+00 5
Coefficients for the Covariates:
beta S.E. CI.lower CI.upper p.value
X1 1.022 0.03004 0.963 1.081 0
X2 3.053 0.02958 2.995 3.111 0
out$est.att
ATT S.E. CI.lower CI.upper p.value n.Treated
-19 0.392159788 0.5468489 -0.6796443 1.46396386 4.732961e-01 0
-18 0.276547958 0.4461493 -0.5978887 1.15098460 5.353532e-01 0
-17 -0.275392926 0.5427657 -1.3391941 0.78840827 6.118824e-01 0
-16 0.441201288 0.4389354 -0.4190963 1.30149888 3.148187e-01 0
-15 -0.889595124 0.4913755 -1.8526735 0.07348322 7.023098e-02 0
-14 0.593890957 0.4633226 -0.3142046 1.50198651 1.999097e-01 0
-13 0.528749012 0.4131945 -0.2810973 1.33859533 2.006643e-01 0
-12 0.171568737 0.5974650 -0.9994412 1.34257868 7.739889e-01 0
-11 0.610832288 0.4474116 -0.2660784 1.48774295 1.721720e-01 0
-10 0.170597468 0.4516495 -0.7146193 1.05581419 7.056379e-01 0
-9 -0.271891657 0.5871305 -1.4226462 0.87886293 6.433030e-01 0
-8 0.094842558 0.5036802 -0.8923526 1.08203768 8.506422e-01 0
-7 -0.651975701 0.5566512 -1.7429921 0.43904067 2.414998e-01 0
-6 0.573686472 0.4782973 -0.3637590 1.51113193 2.303589e-01 0
-5 -0.469685905 0.5150445 -1.4791545 0.53978268 3.618041e-01 0
-4 -0.077766449 0.5287181 -1.1140349 0.95850205 8.830650e-01 0
-3 -0.141784521 0.5472736 -1.2144212 0.93085211 7.955779e-01 0
-2 -0.157100323 0.4186374 -0.9776145 0.66341385 7.074627e-01 0
-1 -0.915575087 0.4855890 -1.8673120 0.03616184 5.936318e-02 0
0 -0.003308833 0.3638145 -0.7163722 0.70975454 9.927435e-01 0
1 1.235962010 0.7289023 -0.1926602 2.66458419 8.995247e-02 5
2 1.630264312 0.5615032 0.5297382 2.73079044 3.691436e-03 5
3 2.712177702 0.5508277 1.6325753 3.79178010 8.486982e-07 5
4 3.466757691 0.7436652 2.0092007 4.92431464 3.135799e-06 5
5 5.740132310 0.5366920 4.6882353 6.79202929 0.000000e+00 5
6 5.280034526 0.5741182 4.1547836 6.40528546 0.000000e+00 5
7 8.436484821 0.4842566 7.4873594 9.38561026 0.000000e+00 5
8 7.839901526 0.6408388 6.5838805 9.09592251 0.000000e+00 5
9 9.455114684 0.5449999 8.3869345 10.52329485 0.000000e+00 5
10 9.638509457 0.5034590 8.6517480 10.62527087 0.000000e+00 5
out$est.avg
Estimate S.E. CI.lower CI.upper p.value
ATT.avg 5.543534 0.262005 5.030014 6.057054 0
out$est.beta
beta S.E. CI.lower CI.upper p.value
X1 1.021890 0.03004240 0.9630082 1.080772 0
X2 3.052994 0.02957771 2.9950231 3.110966 0
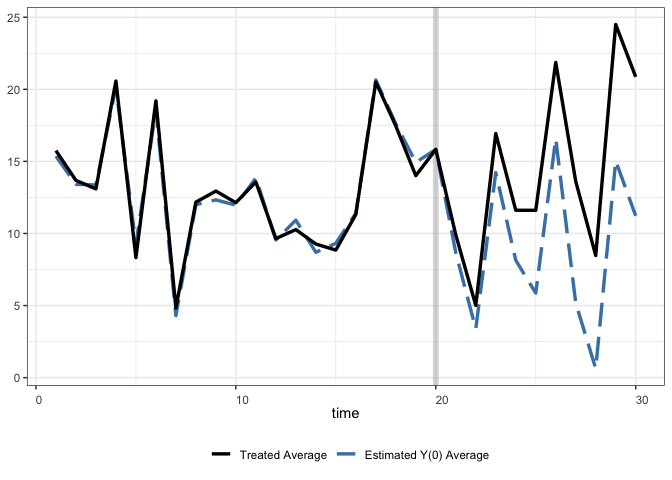
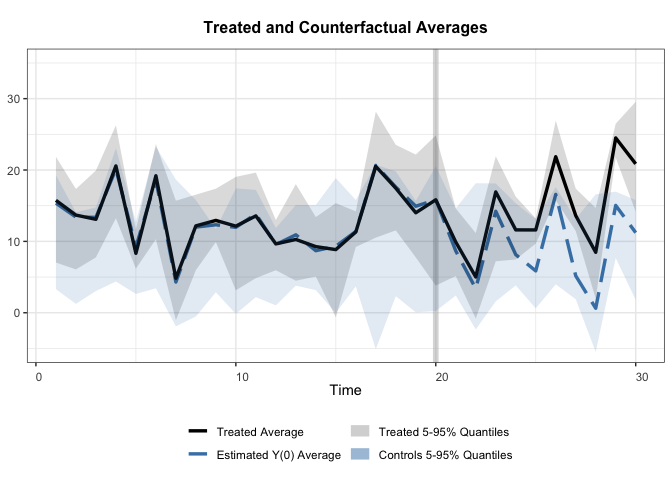
The default plot:
+
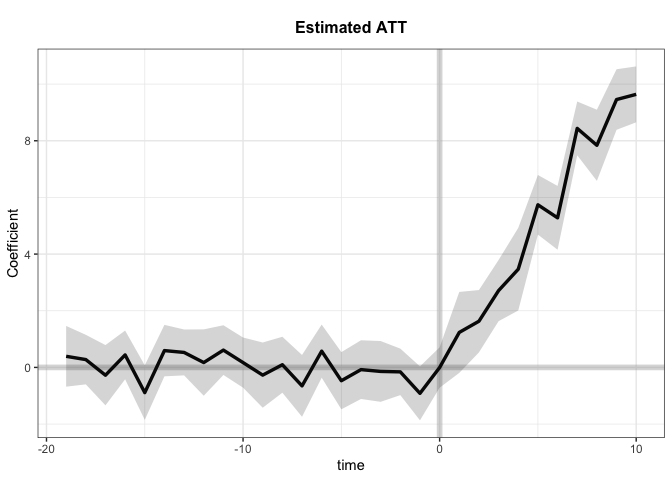
Modified:
+
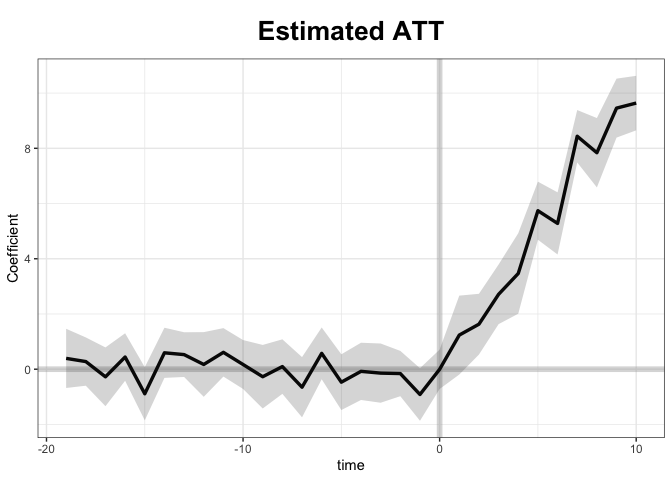
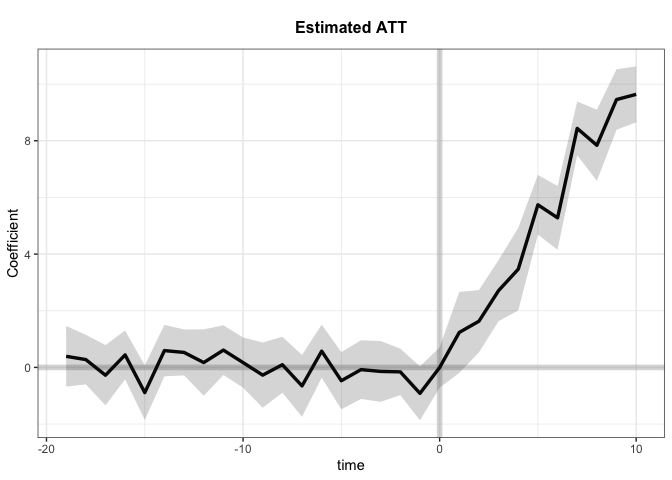
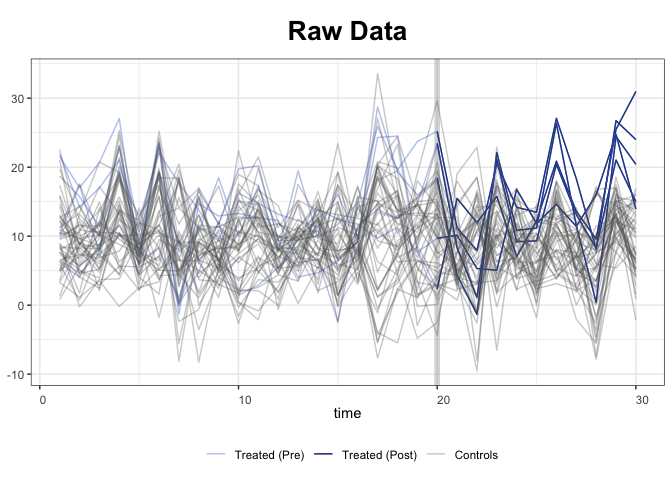
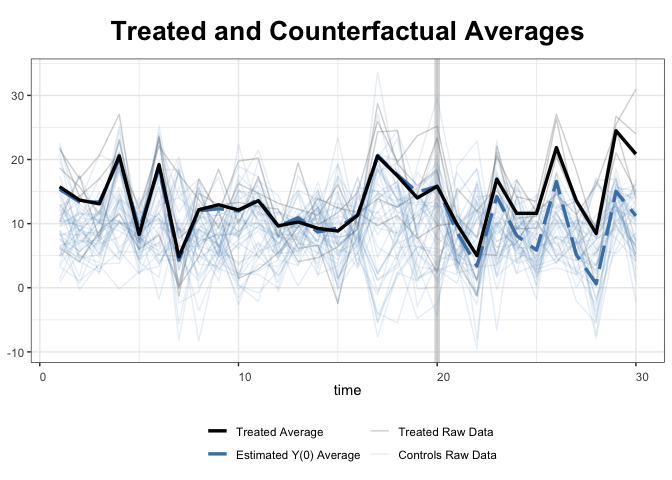
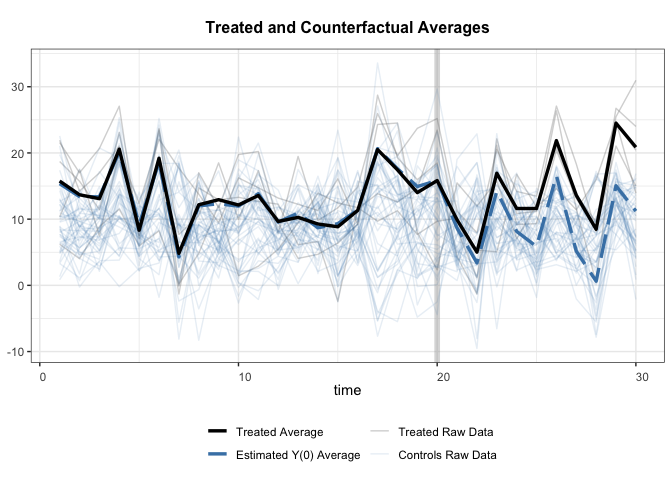
With the raw data:
+
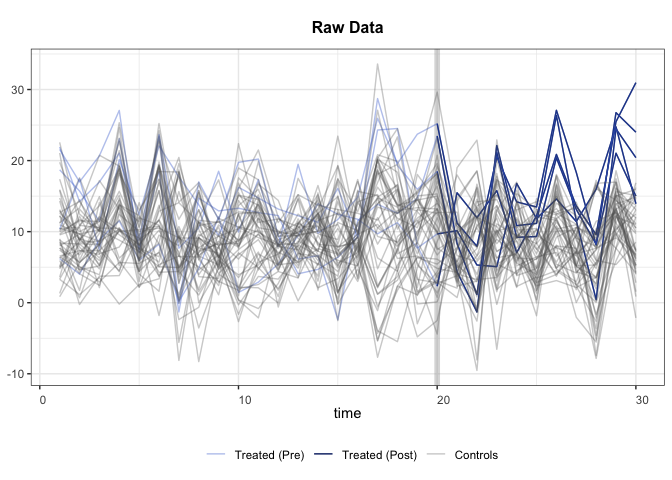
+
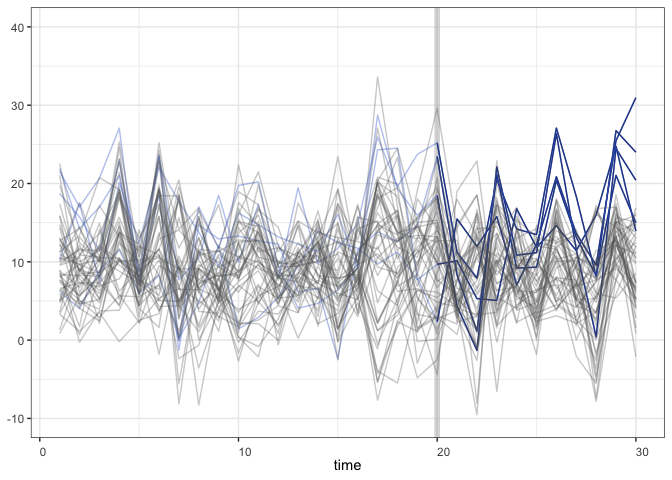
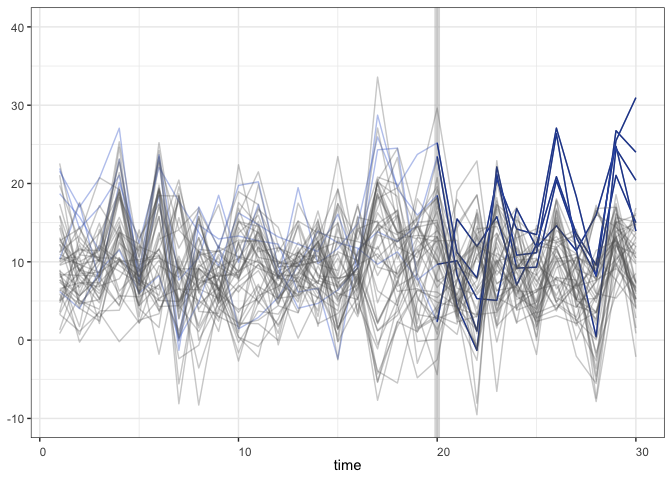
+
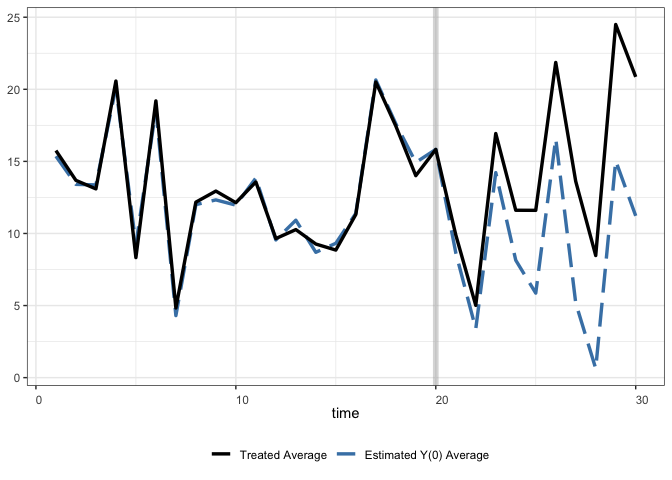
+
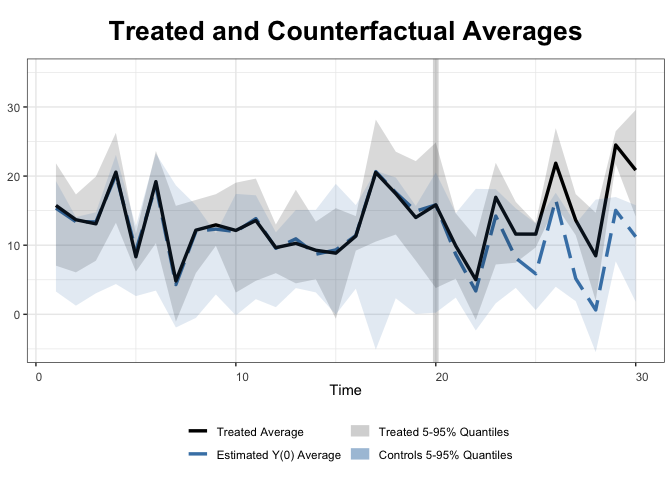
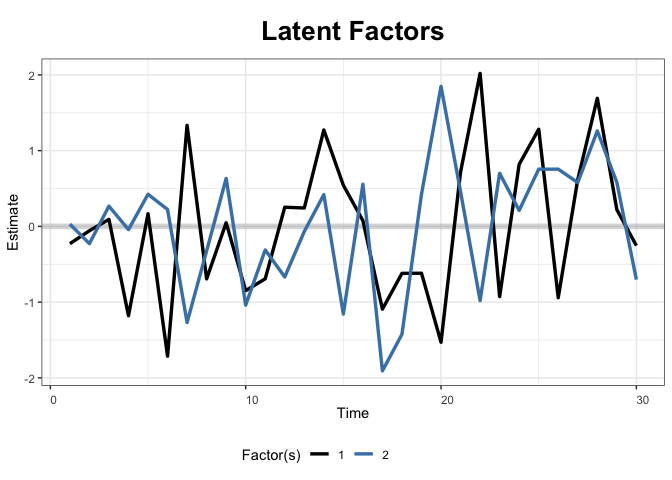
+
+
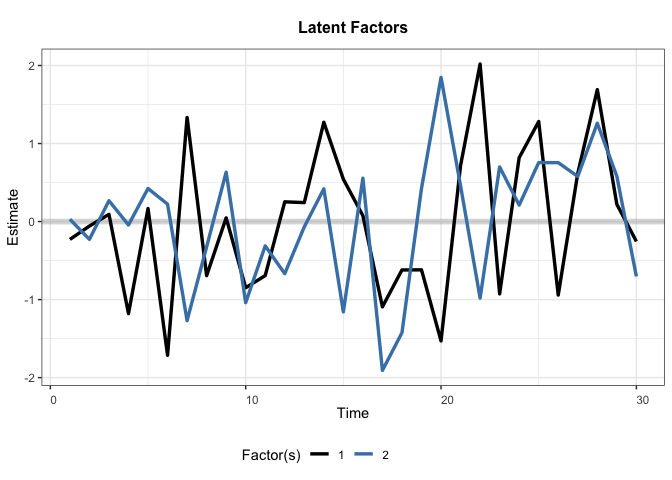
Voter Turnout
Now I am going to try to reproduce the results from Xu’s paper [xu_generalized_2017] and implemented in his package (Xu and Liu 2018).
Data
| abb | year | turnout | policy_edr | policy_mail_in | policy_motor |
|---|---|---|---|---|---|
| AL | 1920 | 21.02107 | 0 | 0 | 0 |
| AL | 1924 | 13.58233 | 0 | 0 | 0 |
| AL | 1928 | 19.03850 | 0 | 0 | 0 |
| AL | 1932 | 17.62002 | 0 | 0 | 0 |
| AL | 1936 | 18.69238 | 0 | 0 | 0 |
| AL | 1940 | 18.86406 | 0 | 0 | 0 |
Model
The paper builds two models, one will only the impact of voter education, another with additional controls. For the sake of time I will build the model with all of the controls used.
out <-
Cross-validating ...
r = 0; sigma2 = 77.99366; IC = 4.82744; PC = 72.60594; MSPE = 22.13889
r = 1; sigma2 = 13.65239; IC = 3.52566; PC = 21.67581; MSPE = 12.03686
r = 2; sigma2 = 8.56312; IC = 3.48518; PC = 19.23841; MSPE = 10.31254*
r = 3; sigma2 = 6.40268; IC = 3.60547; PC = 18.61783; MSPE = 11.48390
r = 4; sigma2 = 4.74273; IC = 3.70145; PC = 16.93707; MSPE = 16.28613
r = 5; sigma2 = 4.02488; IC = 3.91847; PC = 17.05226; MSPE = 15.78683
r* = 2
Simulating errors .............
Bootstrapping ...
..........
The model completed and appeared that it did well. Now to examine the figures to see if they match:
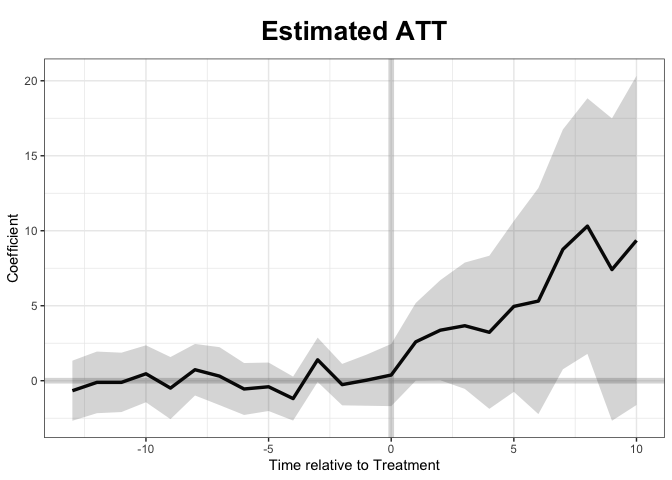
Now we can look at the estimated average treatment
out$est.avg %>%
knitr::
| Estimate | S.E. | CI.lower | CI.upper | p.value | |
|---|---|---|---|---|---|
| ATT.avg | 4.895788 | 2.315232 | 0.3580163 | 9.43356 | 0.0344641 |
+
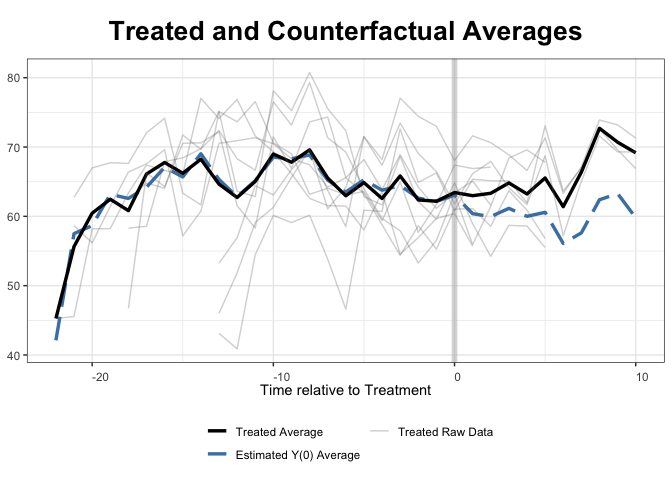
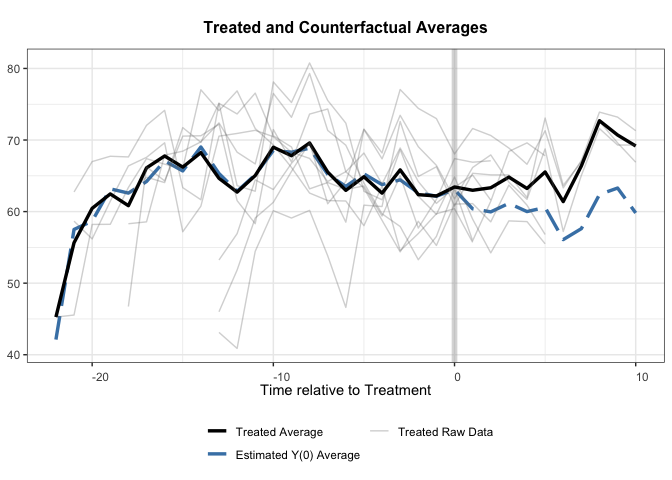
Now with Synth
Now I want to try to replicate the previous case study used in Synth
(Abadie, Diamond, and Hainmueller 2011b) and see if
it is possible.
I am going to recode some variable in order to fit with the methods used
in gsynth. The Synth package uses a dataprep function in order to
format the data into the requirements.
basque_treat <- basque %>%
%>%
knitr::
| regionno | regionname | year | gdpcap | sec.agriculture | sec.energy | sec.industry | sec.construction | sec.services.venta | sec.services.nonventa | school.illit | school.prim | school.med | school.high | school.post.high | popdens | invest | treat |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Spain (Espana) | 1955 | 2.354542 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
| 1 | Spain (Espana) | 1956 | 2.480149 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
| 1 | Spain (Espana) | 1957 | 2.603613 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
| 1 | Spain (Espana) | 1958 | 2.637104 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
| 1 | Spain (Espana) | 1959 | 2.669880 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
| 1 | Spain (Espana) | 1960 | 2.869966 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 |
out_basque <-
Cross-validating ...
r = 0; sigma2 = 0.15833; IC = -1.31080; PC = 0.14556; MSPE = 0.02963
r = 1; sigma2 = 0.02499; IC = -2.64303; PC = 0.04627; MSPE = 0.00994
r = 2; sigma2 = 0.00985; IC = -3.07772; PC = 0.02745; MSPE = 0.00755*
r = 3; sigma2 = 0.00581; IC = -3.12683; PC = 0.02166; MSPE = 0.00981
r = 4; sigma2 = 0.00395; IC = -3.05355; PC = 0.01843; MSPE = 0.00329
r = 5; sigma2 = 0.00256; IC = -3.04564; PC = 0.01435; MSPE = 0.00260
r* = 5
Simulating errors .............
Bootstrapping ...
..........
Let’s check the results
+
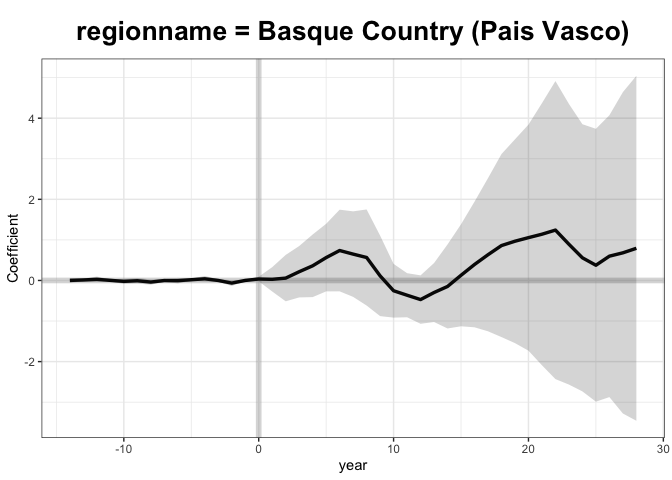
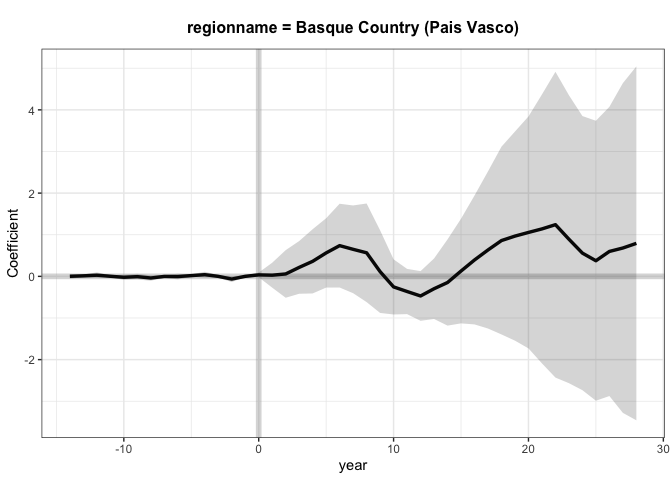
+
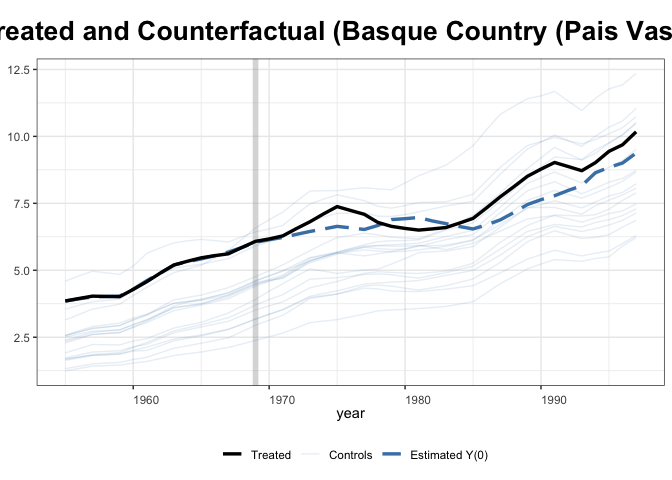
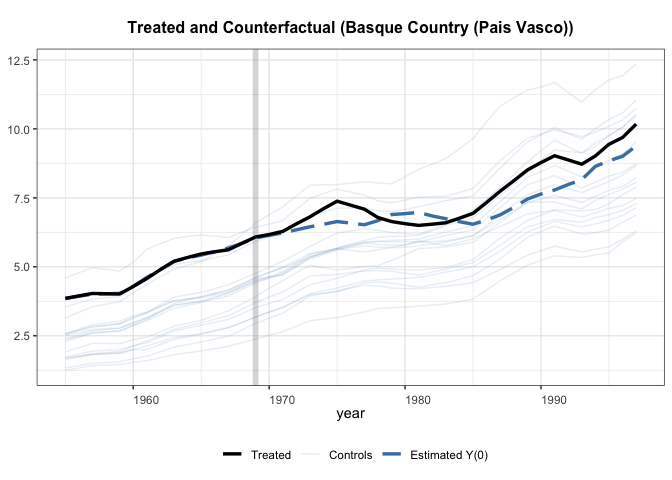
Interesting that this method lands on a slightly different estimate than
the previous paper. I think that this is due to some missing covariates.
It appears that gsynth doesn’t handle missing data too well, which is
ok.
Thoughts
I think that a combination of Bayesian Hierarchical modeling with structural timeseries modeling could get somewhere close to the method, and take advantage of specifying knowns, group effects, etc. But the method is neat and a good way to do some quick analysis.
Abadie, Alberto, Alexis Diamond, and Jens Hainmueller. 2011a. “Synth: An R Package for Synthetic Control Methods in Comparative Case Studies.” Journal of Statistical Software 42 (13): 1–17. http://www.jstatsoft.org/v42/i13/.
———. 2011b. “Synth: An r Package for Synthetic Control Methods in Comparative Case Studies.” Journal of Statistical Software, Articles 42 (13): 1–17. https://doi.org/10.18637/jss.v042.i13.
Xu, Yiqing, and Licheng Liu. 2018. Gsynth: Generalized Synthetic Control Method. https://CRAN.R-project.org/package=gsynth.
Citation
BibTex citation:
@online{dewitt2018
author = {Michael E. DeWitt},
title = {Replicating gsynth},
date = 2018-10-29,
url = {https://michaeldewittjr.com/articles/2018-10-28-replicating-gsynth},
langid = {en}
}
For attribution, please cite this work as:
Michael E. DeWitt. 2018. "Replicating gsynth." October 29, 2018. https://michaeldewittjr.com/articles/2018-10-28-replicating-gsynth